Table information for 'ppakm31.cubes'
General
Table Description:
Spectral cubes of the fields. Look for these with full metadata in
the ivoa.obscore table, obs_collection 'ppakm31 cubes'.
This table is
available for
ADQL queries and through the
TAP endpoint.
Resource Description:
These observations cover five star-forming regions in the Andromeda
galaxy (M31) with optical integral field spectroscopy. Each has a
field of view of roughly 1 kpc across, at 10pc physical resolution. In
addition to the calibrated data cubes, we provide flux maps of the Hβ,
[OIII]5007, Hα, [NII]6583, [SII]6716 and [SII]6730 line emission. Line
fluxes have not been corrected for dust extinction. All data products
have associated error maps.
For a list of all services and tables belonging
to this table's
resource, see Information on resource 'PPAKM31 – Optical Integral Field Spectroscopy of Star-Forming Regions
in M31'
Resource Reference URL:
Resource info
Citing this table
To cite the table as such, we suggest the following
BibTeX entry:
@MISC{vo:ppakm31_cubes,
year=2020,
title={{PPAKM31} – Optical Integral Field Spectroscopy of Star-Forming Regions
in M31},
author={Kreckel, K. and Tomičić, N. and Sandstrom, K. and Groves, B.},
url={http://dc.g-vo.org/tableinfo/ppakm31.cubes},
howpublished={{VO} resource provided by the {GAVO} Data Center}
}
Resource Documentation
For a quick idea of what you can do with this data, consider Kewley
et al's starburst criterion (2001ApJ...556..121K), which
compares [OIII]/Hb with ([SII]6716+[SII]6730)/Ha. In this example, we will
investigate where in the this plane our pixels lie.
We will use pyVO, TOPCAT, and Aladin.
Since we need to do somewhat complex operations on the image pixels,
we'll use pyVO to get the source images and convert them into a table of
fluxes. This table will then be investigated using TOPCAT (and a bit of
Aladin).
Produce a flux table: You could use Aladin for going from the images to a
table for fluxes, but this is a bit of drudgery here. Instead, here is a short
python script using pyVO and numpy; it uses TAP to retrieve the image metadata
and then get the images themselves by plain HTTP.
It then filters out all pixels with an SNR below 5 (with a bit of handwaving,
assuming what's ok in the generally weakest band will be ok in the others)
puts the remaining pixels into a nice relational table. It finally sends
this table to TOPCAT, so start TOPCAT before running this:
from astropy.io import fits as pyfits
from astropy import table, wcs
from pyvo import samp
import numpy
import pyvo
# change the following to use other ppakm31 fields
FIELD = "F3"
def stringify(s):
"""returns s utf-8-decoded if it's bytes, s otherwise.
(that's working around old astropy votable breakage)
"""
if isinstance(s, bytes):
return s.decode("utf-8")
return s
def get_image_metadata(field):
"""returns metadata rows from the ppakm31 service for field.
Access URL and schema are taken from a TOPCAT exploration of the service.
"""
svc = pyvo.dal.TAPService("http://dc.g-vo.org/tap")
res = []
for row in svc.run_sync(
"select accref, field, imagetitle, bandpassid, cube_link"
" from ppakm31.maps"
" where field={}".format(repr(field))):
res.append({
"bandpassid": stringify(row["bandpassid"]),
"accref": stringify(row["accref"]),})
return res
def get_line_maps(image_meta):
"""returns a dict band->hdus for image_meta as returned by
get_image_maps.
"""
res = {}
for row in image_meta:
if row["bandpassid"]=='[NII]6583':
# we don't need that one later, so skip it
continue
res[row["bandpassid"]] = pyfits.open(row["accref"])
return res
if __name__=="__main__":
line_maps = get_line_maps(get_image_metadata(FIELD))
# Select pixels with a reasonable SNR; the errors are in extension
# 1, and we use [OIII]5007, since the other lines ought to be stronger
snr_cutoff = 5.
snr_mask = (line_maps["[OIII]5007"][0].data
> line_maps["[OIII]5007"][1].data*snr_cutoff)
# The line maps all have identical WCS: use any to turn pixels to pos.
# This is a somewhat tricky way to get the positions of all valid
# pixels:
w = wcs.WCS(header=line_maps["Halpha"][0].header, naxis=2)
ras, decs = w.wcs_pix2world(
snr_mask.nonzero()[1], snr_mask.nonzero()[0], 1)
# Create table with ra & dec positions and our flux measurements.
t = table.Table([
ras,
decs,
line_maps["[OIII]5007"][0].data[snr_mask],
line_maps["Hbeta"][0].data[snr_mask],
line_maps["Halpha"][0].data[snr_mask],
line_maps["[SII]6716"][0].data[snr_mask],
line_maps["[SII]6730"][0].data[snr_mask]],
names=('ra', 'dec', 'OIII_5007','Hbeta','Halpha','SII_6716','SII_6730'))
# Send the table to TOPCAT via SAMP
with samp.connection() as conn:
samp.send_table_to(conn, t, "topcat")
You can change FIELD (or change the script so your flux table has all fields).
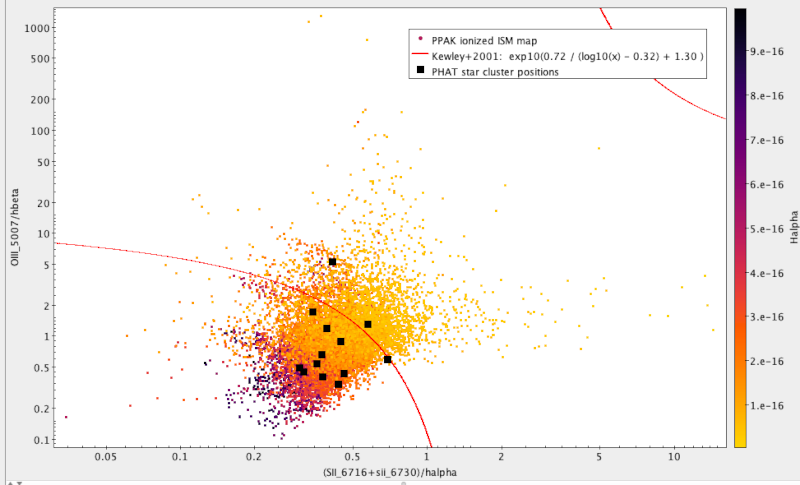
Make a plot of the line ratios: To visualise where the data likes
with respect to the Kewley+2001 criterion, once you have the table in TOPCAT,
configure the plot rougly likes this:
- Graphics → Plane Plot
- In X, type the denominator of the Kewley criterion; in case you forget how
the columns are called, check Views → Column Info. Anyway, it's
(SII_6716+SII_6730)/Halpha
- Similarly, for Y, use OIII_5007/Hbeta.
- In the Form tab, set the shading mode to aux, and set the Aux axis to
Halpha
- Under Axes, set both axes to log.
- Now add the Kewley criterion with Layers → Add Function Control and adding
using exp10(0.72/(log10(x)-0.32)+1.30) as the function to plot; to see
where this comes from, read 2001ApJ...556..121K.
The result would be something like this (see below for the black squares):
Sanity check: To see how the various points on your plot look in reality,
start Aladin and tell it to either some common survey or PPAKM31 images
themselves; you can find the latter in the discovery tree left in Aladin's
window.
Then, configure TOPCAT can make Aladin follow its focus by clicking Views →
Activation Action and then checking “Send Sky Coordinates”. See if you can
correlate the position in the plot with the visual (or infrared, or whatever)
appearance.
Compare to known clusters: To see what known star clusters fare in our plot,
get a catalogue of them. In TOPCAT, use VO → Cone Search, and in keywords,
enter something like “M31 cluster”; to find what we've plotted above,
J/ApJ/827/33, you'll have to check “description” in the match fields before
sending off the query (they don't mention M31 in their title or subject); but,
really, other catalogues of clusters or H II regions should do just as well.
To quickly get a position to search for, click into your plot; this will fill
out RA and Dec in the dialog, and all that's left is to enter, perhaps, 0.1 into
the radius field (because that's about the FoV of our images) and fire off the
request.
Whatever catalogue you used, the entries will probably not have the fluxes we
need to put them into our flux ratio plot. So: let's just add
them from our data by a positional crossmatch. To do that, in TOPCAT's main
window say Joins → Pair Match. Our pixel size is about 1 arcsec, so the default
match radius is ok. Select our fluxes as table 1 and the cluster catalogue as
table 2, hit “Go”, and then return to the flux plot.
In there, do Layers → Add position control. Configure this to plot the match
table, and again the flux expressions from above. If you then tell TOPCAT to
plot the points as large black squares, you will have reproduced (more or less)
the plot above.
Columns
Sorted by DB column index. [Sort alphabetically]
| Name | Table Head |
Description | Unit | UCD |
|---|
| accref |
Product key |
Access key for the data
|
N/A |
meta.ref.url |
| owner |
Owner |
Owner of the data
|
N/A |
N/A |
| embargo |
Embargo ends |
Date the data will become/became public
|
yr |
N/A |
| mime |
Type |
MIME type of the file served
|
N/A |
meta.code.mime |
| accsize |
File size |
Size of the data in bytes
|
byte |
VOX:Image_FileSize |
| obs_id |
Obs. Id |
Unique identifier for an observation
|
N/A |
meta.id |
| obs_publisher_did |
Pub. DID |
Dataset identifier assigned by the publisher.
|
N/A |
meta.ref.ivoid |
| access_estsize |
Size |
Estimated size of data product
|
kbyte |
phys.size;meta.file |
| obs_title |
Title |
Free-from title of the data set
|
N/A |
meta.title;obs |
| s_ra |
RA |
RA of (center of) observation, ICRS
|
deg |
pos.eq.ra |
| s_dec |
Dec |
Dec of (center of) observation, ICRS
|
deg |
pos.eq.dec |
| t_min |
Min. t |
Lower bound of times represented in the data set
|
d |
time.start;obs.exposure |
| t_max |
Max. t |
Upper bound of times represented in the data set
|
d |
time.end;obs.exposure |
| s_region |
coverage |
Region covered by the observation, as a polygon
|
N/A |
pos.outline;obs.field |
| em_min |
λ_min |
Minimal wavelength represented within the data set
|
m |
em.wl;stat.min |
| em_max |
λ_max |
Maximal wavelength represented within the data set
|
m |
em.wl;stat.max |
| s_xel1 |
|X| |
Number of elements (typically pixels) along the first spatial axis.
|
N/A |
meta.number |
| s_xel2 |
|Y| |
Number of elements (typically pixels) along the second spatial axis.
|
N/A |
meta.number |
Columns that are parts of indices are marked
like this.
Other
The following services may use the data contained in this
table:
VOResource
VO nerds may sometimes need
VOResource XML for this table.
![[Operator logo]](/static/img/logo_medium.png)
